Principal Investigators
Attila Gürsoy
Research Area
Özlem Keskin
Research
For more information see Dr. Keskin’s homepage.
PhD Students
-
Damla ÖvekPhD Candidate, Computer Science and Engineering

-
Meryem ErenPhD Candidate, Molecular Biology and Genetics

-
Erxiati PaerhatiPhD Student, Computational Science and Engineering

-
Melisa Ece ZeylanPhD Student, Computational Science and Engineering

-
Fatma CankaraPhD Candidate, Computational Science and Engineering

-
Esra BaşaranPhD Student, Computational Science and Engineering

-
Simge ŞenyüzPhD Student, Computational Science and Engineering

-
Kayra KösoğluPhD Student, Computational Science and Engineering
-
Zeynep AbalıPhD Student, Computational Science and Engineering

-
Zeynep AydınPhD Student, Computational Science and Engineering

-
Amir FathiPh.D. Student, Computational Science and Engineering

Damla Övek
Research
Short Bio
Ph.D. Candidate, AI Fellow, Koç University-Computer Science & Engineering
M.Sc. Koç University-Computational Science & Engineering
B.Sc. Koç University-Computer Engineering (Double Major)
B.Sc. Koç University-Molecular Biology & Genetics
Meryem Eren
Research
Short Bio
Erxiati Paerhati
Research
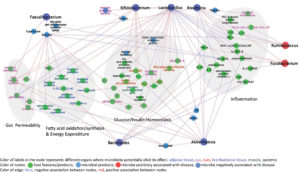
Investigation of the association between type2 diabetes and human gut microbiota
Short Bio
Ph.D. Student Koç University-Computational Science and Engineering
B.Sc. Hacettepe University-Biology
Melisa Ece Zeylan
Research Area
Cardiovascular stress impacts on neuronal function: intracellular pathways to cognitive impairment
Short Bio
Fatma Cankara
Research
Short Bio
Esra Başaran
Research
Short Bio
Simge Şenyüz
Short Bio
Ph.D. Candidate, Koç University, Computational Science and Engineering
M.Sc. Koç University, Computational Science and Engineering
B.Sc. Boğaziçi University, Chemistry
M.Sc Thesis: The Investigation of Mechanistic Differences of Rac1P29S and Rac1A159V Activation via Molecular Dynamics Simulations
Research
Cardiovascular stress impacts on neuronal function: intracellular pathways to cognitive impairment
Kayra Kösoğlu
Research
M.Sc. Thesis: Turning the Spotlight on a Long-Overlooked Pathway: Molecular Dynamics Investigation of Ras Family Members with RalGDS.
Short Bio
Ph.D. Candidate Koç University, Computational Science and Engineering
M.Sc. Koç University, Molecular Biology and Genetics
B.Sc. Istanbul Bilgi University, Genetics and Bio-engineering
Zeynep Abalı
Ph.D. Student, AI Fellow, Koç University – Computational Science and Engineering M.Sc. Thesis:Short Bio
M.Sc. Koç University, Data Science
B.Sc. Middle East Technical University – Molecular Biology and Genetics
A Data-Centric Approach for Investigation of Protein-Protein Interfaces in Protein Data Bank
Drug Repurposing Using Structural Similarity and Discovery of New Target ProteinsResearch
Zeynep Aydın
Ph.D. Candidate, Koç University, Computational Science and Engineering B.Sc. Short Bio
Research
Amir Fathi
Ph.D. Candidate, Koç University, Computational Science and EngineeringShort Bio
M.Sc.
B.Sc.
Research
MSc Students
-
Moaaz KhokharM.Sc. Student, Computer Science and Engineering

-
Zeynep Ahenk SayınM.Sc. Student, Chemical and Biological Engineering

Moaaz Khokhar
M.Sc. Candidate, AI Fellow, Koç University, Computer Science and EngineeringShort Bio
B.Sc.
Research
Zeynep Ahenk Sayın
M.Sc. Candidate Koç University, Chemical and Biological EngineeringShort Bio
B.Sc.
Ras/Raf/MAPK Pathway Structural NetworkResearch
Undergrad Students
-
Demet TümkayaUndergrad DM Student, Chemical and Biological Engineering & Computer Engineering

-
Yiğit Can AteşUndergrad DM Student, Molecular Biology and Genetics & Computer Engineering

-
Tolga ErmişUndergrad DM Student, Chemical and Biological Engineering & Computer Engineering

Demet Tümkaya
Research
Assessment of PRISM Software by using Apoptosis Pathway

Short Bio
B.Sc. Koç University – Chemical-Biological Engineering & Computer Engineering
Yiğit Can Ateş
Short Bio
Tolga Ermiş
Short Bio
B.Sc. Koç University – Chemical-Biological Engineering & Computer Engineering
Alumni
Recent
-
Emre KüçükMSc, Data Science

-
Burcu ÖzerBSc, Chemical and Biological Engineering & Computer Engineering (DM)

-
Jackson WeakoPhD, Computational Science and Engineering

-
Sukejna ValjevacMSc, Computer Science and Engineering

-
Ali Tuğrul BalcıMSc, Computer Science and Engineering

-
Ayşe Seda YazgılıMSc, Molecular Biology and Genetics

-
Emine Sıla ÖzdemirPhD, Chemical and Biological Engineering

-
Serena MuratcıoğluPhD, Chemical and Biological Engineering

-
Efe ElbeyliMSc, Biomedical Science and Engineering

-
Bilgesu ErdoğanMSc, Biomedical Science and Engineering

-
Asma HakouzMSc, Computer Science and Engineering

-
Meltem Eda ÖmürMSc, Molecular Biology and Genetics

-
Farideh HalakouPhD, Computer Science and Engineering

Emre Küçük
Short Bio
M.Sc. Student Koç University-Data Science
B.Sc. Istanbul Technical University-Mathematical Engineering
Burcu Özer
Research
Assessment of PRISM Software by using Apoptosis Pathway

Short Bio
B.Sc. Koç University – Chemical-Biological Engineering & Computer Engineering
Jackson Weako
Research
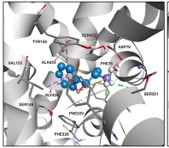
Understanding the Pleckstrin Homology (PH) Domain Peculiar Mechanism in Akt Translocation, Phosphorylation, and Activation.
Short Bio
Ph.D. Koç University, Computational Science and Engineering
M.Sc. Kadir Has University, Computational Biology and Bioinformatics
B.Sc. Cuttington University, Chemistry
For more information see Jackson’s homepage
Sukejna Valjevac
Research
Evaluation and Ranking of Protein Docking Models by 3D Convolutional
Neural Networks
Short Bio
M.Sc. Koç University-Computer Science & Engineering
B.Sc. Boğaziçi University-Molecular Biology &Genetics
Ali Tuğrul Balcı
Research
Since proteins are pivotal for the most of the crucial activities that are happening in organisms and the proteins, most of the time, do not act alone, studying protein-protein interactions is very important. Predicting which proteins can interact with each other is a very valuable skill on which I am working by using both statistical and deterministic algorithms.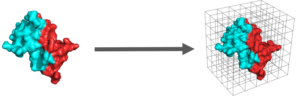
Short Bio
M.Sc. Koç University-Computer Science and Engineering
Ayşe Seda Yazgılı
Research
Ras protein is shown to interact with the proteins involved in crucial pathways 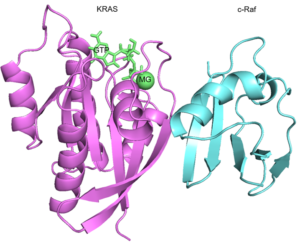 such as MAPK and PI3K. Our group recently showed that specific mutations on Ras protein interfere with the p-ERK level within the cell. Using molecular dynamics simulation methods and computational analysis, we aim to have a deeper understanding how these mutations affects the interaction .Therefore, I am simulating the interactions of KRAS protein and Ras-Binding-Domain (RBD) of c-Raf protein.
such as MAPK and PI3K. Our group recently showed that specific mutations on Ras protein interfere with the p-ERK level within the cell. Using molecular dynamics simulation methods and computational analysis, we aim to have a deeper understanding how these mutations affects the interaction .Therefore, I am simulating the interactions of KRAS protein and Ras-Binding-Domain (RBD) of c-Raf protein.
Short Bio
M.Sc. Koç University-Molecular Biology and Genetics
B.Sc. İstanbul Technical University-Molecular Biology and Genetics
For more information see Ayşe Seda’s homepage
Emine Sıla Özdemir
Research
IQGAP2 is a scaffold protein which plays central roles in cell-cell  adhesion, polarity and cell motility. Rho GTPases Cdc42 and Rac1, in their GTP-bound active forms, interact with all three human IQGAP isoforms. IQGAP-Cdc42 interaction promotes metastasis by enhancing actin polymerization. However, despite their high degree of sequence identity, Cdc42 and Ra1 intrections with IQGAP show differences. We aim to reveal the interaction mechanisms of these proteins.
adhesion, polarity and cell motility. Rho GTPases Cdc42 and Rac1, in their GTP-bound active forms, interact with all three human IQGAP isoforms. IQGAP-Cdc42 interaction promotes metastasis by enhancing actin polymerization. However, despite their high degree of sequence identity, Cdc42 and Ra1 intrections with IQGAP show differences. We aim to reveal the interaction mechanisms of these proteins.
Publications
Unraveling the molecular mechanism of interactions of the Rho GTPases Cdc42 and Rac1 with the scaffolding protein IQGAP2. Ozdemir ES, Jang H, Gursoy A, Keskin O, Li Z, Sacks DB, Nussinov R (2018) . The Journal of Biological Chemistry. doi:10.1074/jbc.RA117.001596
Relation between Protein Intrinsic Normal Mode Weights and Pre-Existing Conformer Populations. Ozgur B*, Ozdemir ES*, Gursoy A, Keskin O (2017) Journal of Physical Chemistry B 121 (15):3686-3700.doi:10.1021/
Red emitting, cucurbituril-capped, pH-responsive conjugated oligomer-based nanoparticles for drug delivery and cellular imaging. Pennakalathil J, Jahja E, Ozdemir ES, Konu O, Tuncel D (2014) Biomacromolecules 15 (9):3366-3374. doi:10.1021/bm500839j
Short Bio
M.Sc. Bilkent University-Molecular Biology and Genetics
Ph.D. Koç University-Chemical and Biological Engineering
For more information see Sıla’s homepage
Serena Muratcıoğlu
Research
Ras proteins recruit and activate effectors, including Raf, that transmit receptor-initiated signals and regulates gene expression. Ras is believed to function as a monomer; however, since Raf dimerizes, it was suspected that Ras may also dimerize. Here w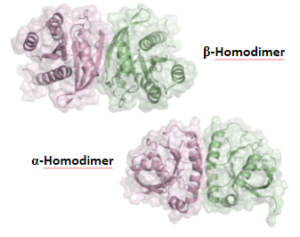 e show that GTP-bound K-Ras4B forms stable homodimers using two interfaces. We aim to confirm RAS dimerization in vivo and understand the effect of
e show that GTP-bound K-Ras4B forms stable homodimers using two interfaces. We aim to confirm RAS dimerization in vivo and understand the effect of
Short Bio
Ph.D. Koç University-Chemical and Biological Engineering
B.Sc. Istanbul Technical University-Chemistry and Molecular Biology & Genetics
Efe Elbeyli
Research
Capri is community wide experiment to test protein-protein docking methods  in blind predictions based on three-dimensional structures. Targets are selected from experimentally known but unpublished complex structures. Predictors can use any of the information in literature about the components for prediction.
in blind predictions based on three-dimensional structures. Targets are selected from experimentally known but unpublished complex structures. Predictors can use any of the information in literature about the components for prediction.
Short Bio
M.Sc. Koç University-Biomedical Science and Engineering
Bilgesu Erdoğan
Research
Diseases are commonly the result of dysregulated complex interactions involving large sets of genes and proteins as products of these genes, and their cooperation with other cellular components. Interpreting protein-protein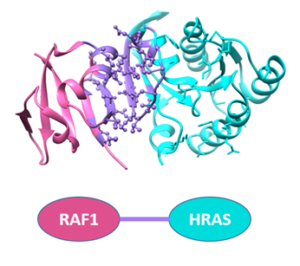 interactions at both network and molecular interaction levels with mutation knowledge requires a comprehensive research process that is fed from different sources. In this project, we developed a web-based tool, Gene2Phen, by integrating large-scale protein-protein interaction network, 3D protein structure information and interface mutation knowledge to aid researchers in exploring and comparing the molecular mechanism of different phenotypes. Gene2Phen works as an automatized pipeline tool to build, visualize and compare phenotype specific subnetworks, to examine protein- protein interactions associated with their structure and mutation data. Gene2Phen web tool prioritizes the human protein-protein network based on seed genes specific to a phenotype. From the prioritized-PPI
interactions at both network and molecular interaction levels with mutation knowledge requires a comprehensive research process that is fed from different sources. In this project, we developed a web-based tool, Gene2Phen, by integrating large-scale protein-protein interaction network, 3D protein structure information and interface mutation knowledge to aid researchers in exploring and comparing the molecular mechanism of different phenotypes. Gene2Phen works as an automatized pipeline tool to build, visualize and compare phenotype specific subnetworks, to examine protein- protein interactions associated with their structure and mutation data. Gene2Phen web tool prioritizes the human protein-protein network based on seed genes specific to a phenotype. From the prioritized-PPI  network, users can generate a phenotype specific subnetwork. The phenotype-specific subnetworks can be visualized and compared interactively. Genome annotations and topological properties of each protein are shown in this interactive network representation. A unique feature of Gene2Phen is its ability to display 3D structural models of protein-protein interactions and their predicted protein-protein interfaces. Users can see the list of mutations which are mapped on predicted protein-protein interfaces. This allows users to study mutations altering protein-protein interfaces and their role in the phenotype-specific subnetworks. Gene2Phen, by automating the integration of protein-protein networks, protein structure, and disease – related mutations at large scale, will not only boost the productivity and efficiency, but it may be the leveraging step to the novel solutions/studies.
network, users can generate a phenotype specific subnetwork. The phenotype-specific subnetworks can be visualized and compared interactively. Genome annotations and topological properties of each protein are shown in this interactive network representation. A unique feature of Gene2Phen is its ability to display 3D structural models of protein-protein interactions and their predicted protein-protein interfaces. Users can see the list of mutations which are mapped on predicted protein-protein interfaces. This allows users to study mutations altering protein-protein interfaces and their role in the phenotype-specific subnetworks. Gene2Phen, by automating the integration of protein-protein networks, protein structure, and disease – related mutations at large scale, will not only boost the productivity and efficiency, but it may be the leveraging step to the novel solutions/studies. 
Short Bio
M.Sc. Koç University-Biomedical Science and Engineering
B.Sc. Bilkent University-Computer Engineering
Asma Hakouz
Research
Short Bio
M.Sc. Koç University – Computer Science and Engineering
B.Sc. The University of Jordan – Electrical Engineering
For more information see Asma’s homepage
Meltem Eda Ömür
Research
Classification of structures of protein-protein interactions may provide 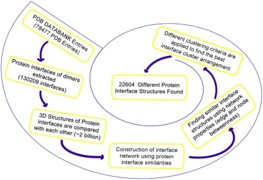 valuable insights for modeling and abstracting design principles. We aim to cluster protein-protein interactions by their interface structures, and to exploit these clusters to obtain and study shared and distinct protein binding sites and expand previously created interface datasets. The novelty in this work is in generating structurally non-redundant protein-protein interfaces which are sensitive to small perturbations in protein binding sites that have a significant impact in template-based docking.
valuable insights for modeling and abstracting design principles. We aim to cluster protein-protein interactions by their interface structures, and to exploit these clusters to obtain and study shared and distinct protein binding sites and expand previously created interface datasets. The novelty in this work is in generating structurally non-redundant protein-protein interfaces which are sensitive to small perturbations in protein binding sites that have a significant impact in template-based docking.
Short Bio
M.Sc. Koç University-Molecular Biology and Genetics
B.Sc. Üsküdar University-Computer Engineering (Double Major)
B.Sc. Üsküdar University-Molecular Biology and Genetics
Farideh Halakou
Research
The common practice in structural PPI networks is to consider just one specific conformation for each protein. Yet it is not a comprehensive represent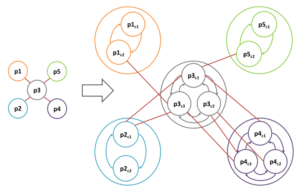 ation as it neglects the conformational changes of proteins which may lead to different protein interactions, functions, and downstream signaling. We propose a new representation for structural PPI networks which inspects the different conformations of proteins.
ation as it neglects the conformational changes of proteins which may lead to different protein interactions, functions, and downstream signaling. We propose a new representation for structural PPI networks which inspects the different conformations of proteins.
Publications
Enriching Traditional Protein-protein Interaction Networks with Alternative Conformations of Proteins. Halakou F, Kilic ES, Cukuroglu E, Keskin O, Gursoy A. Sci Rep. 2017 Aug 3;7(1):7180.
Topological, functional, and structural analyses of protein-protein interaction networks of breast cancer lung and brain metastases, Halakou F, Gursoy A., Kilic E. S. and Keskin O. 2017 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Manchester, 2017, pp. 1-7.
doi: 10.1109/CIBCB.2017.8058539
Short Bio
M.Sc. Kerman Graduate University of Technology-Information Technology Engineering
For more information see Farideh’s homepage

