Most biological processes involve interactions between proteins. Characteristics of the interaction and interaction partners determine the downstream processes, and perturbations of such interactions are associated with various human diseases.
Proteins interact with other proteins or molecules through protein interfaces (PPIs). Drugs or small molecules can block or alter the activity of the proteins by binding to these sites. Due to this susceptibility to modulation of PPIs, they constitute a highly promising type of potential target for pharmacological intervention. The discovery of potential drug targets is a challenge for drug-development applications owing to the complexity and variety of targeted regions. Thus characterizing PPIs and understanding their disruption mechanisms help us to develop effective strategies to tackle labor-intensive drug discovery and repurposing procedures. Computational applications are taking a bigger part in the analysis of protein-protein interactions and by better understanding these interfaces, we can discover new possible target proteins for drugs that are already in use.
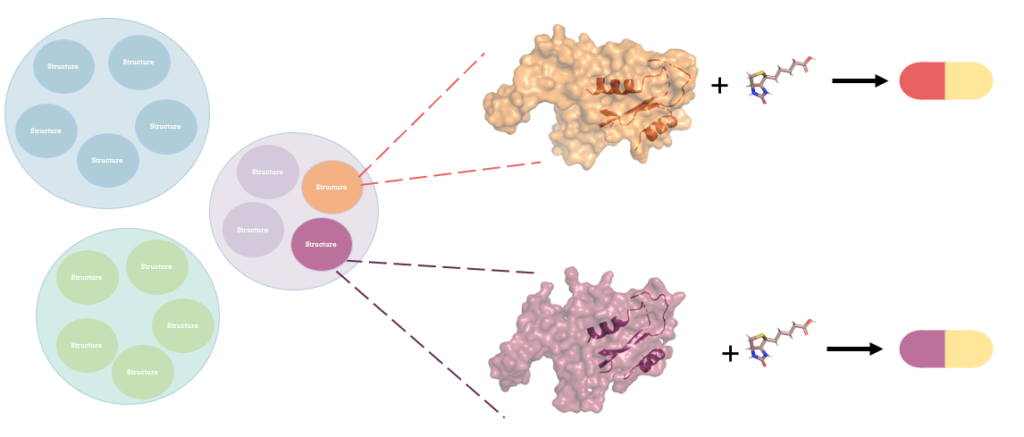
Drug repurposing is a strategy for finding new targets for the existing approved or investigational drugs that are not initially considered targets in the drug design process. This is an advantageous strategy given the labor and financial cost of developing a new drug. Additionally, given that selected drugs are approved before or at least found safe to use in humans, this strategy vastly eliminates the toxicity effects of the repurposed drugs. The key idea here is that similar drugs target similar proteins.
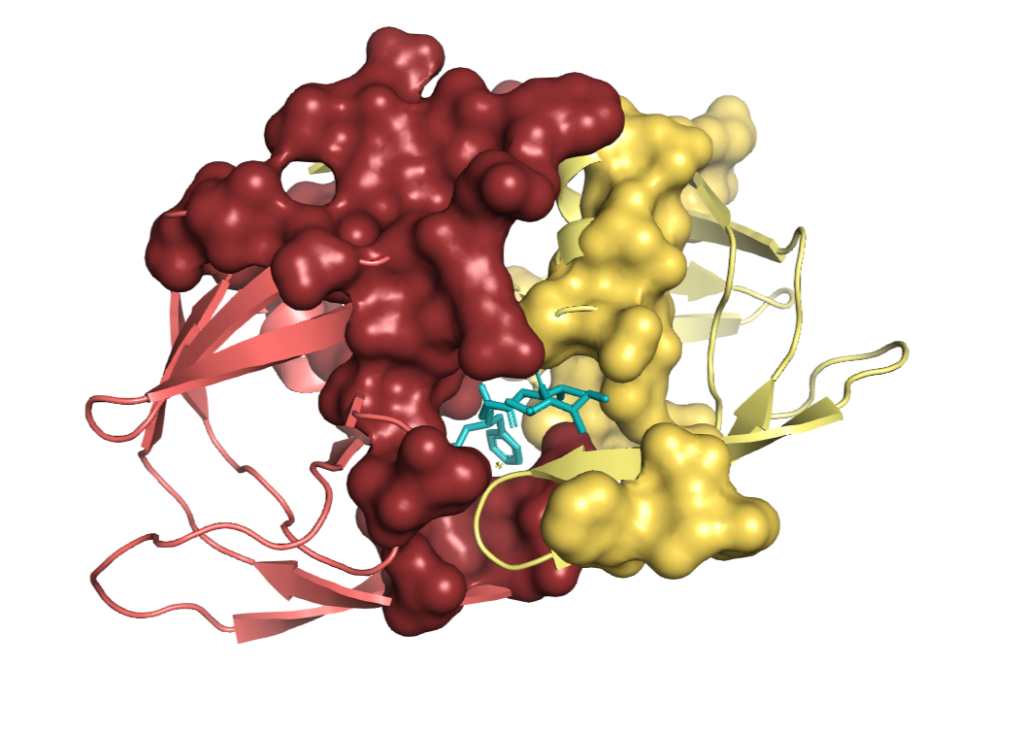
In this figure, purple and orange circles representing two proteins belong to the same cluster, thus expected to have similar interface regions. Cartoon representation inside surface representation of proteins indicates interface regions. Since they belong to the same cluster, both contain two alpha-helices and two beta-sheets. Ligand shown in the figure thus is expected to interact with both interface regions. As a result of these interactions, a drug that was designed for the interaction of orange protein can be repurposed to interact with the purple protein and be therapeutically effective on another pathway.
In our group, we aim to use this strategy and facilitate drug targeting of PPIs by means of structural clustering of interface regions and drug-like ligands. Structural clustering of interface regions allows the detection of structurally similar interaction regions. Hypothetically, a ligand that binds to an interface can bind to another similar interface of another protein from the same cluster.
Our hypothesis is that if we can identify similar interfaces and similar drugs, search space for drug repurposing studies will be decreased dramatically and this in turn will increase the chances of hitting the correct target. To achieve this, we apply a strategy to group protein interfaces according to the structural similarity.
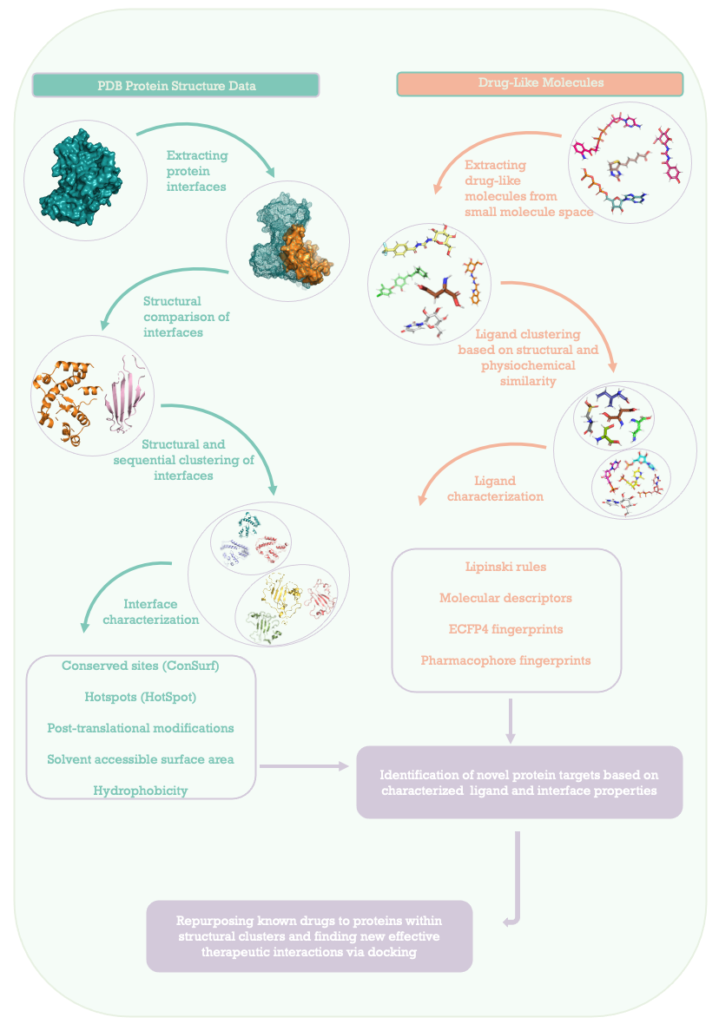
- We created a protein-protein interface database using the 3D structures of the protein-protein complexes from Protein Data Bank. This database allows the identification of structurally similar interface regions, and thus facilitates the search for alternative targets.
- We retrieved drug-like molecules from different websites and generated a dataset of candidate ligand molecules for the repurposing study.
- To characterize drug-like molecules, we evaluated a number of physical and chemical properties that will allow us to identify molecules that show a greater tendency to interact with interfaces whose drug-like partners are known and similar to the molecule of interest.
- Along with such properties, we also clustered ligand molecules using ECFP4 and pharmacophore fingerprints and obtained ligand clusters that will help us identify similar drug-like molecules.
- We used molecular approximation approach to test possible drug-protein interactions and determine new target proteins for the drug-like small molecules.
- By doing so, we aim to identify similar interfaces that can be targeted by alternative drugs. This approach will help to enlarge the available drug space for proteins and increase the likelihood of hitting the correct target.
Project Members
Funded By:
Türkiye Sağlık Enstitüleri Başkanlığı
