At the start of the human genome project, estimated number of genes in the human genome was around 100.000. When the initial draft genome was announced, it was realized that the total number of genes were much lower than the estimations, around ~20.000.[1] This discovery supported the view that the biological complexity cannot be fully explained with the number of genes a species possesses and brought up a new corridor for research to explain the high complexity of humans at the organismal level compared with other organisms who exhibited lower complexity but similar gene numbers.
With extensive computational and experimental effort, contribution of several mechanisms to the complexity observed in molecular and organismal level have been appreciated, these include alternative splicing, post-translational modifications, and diversity of protein-protein interactions
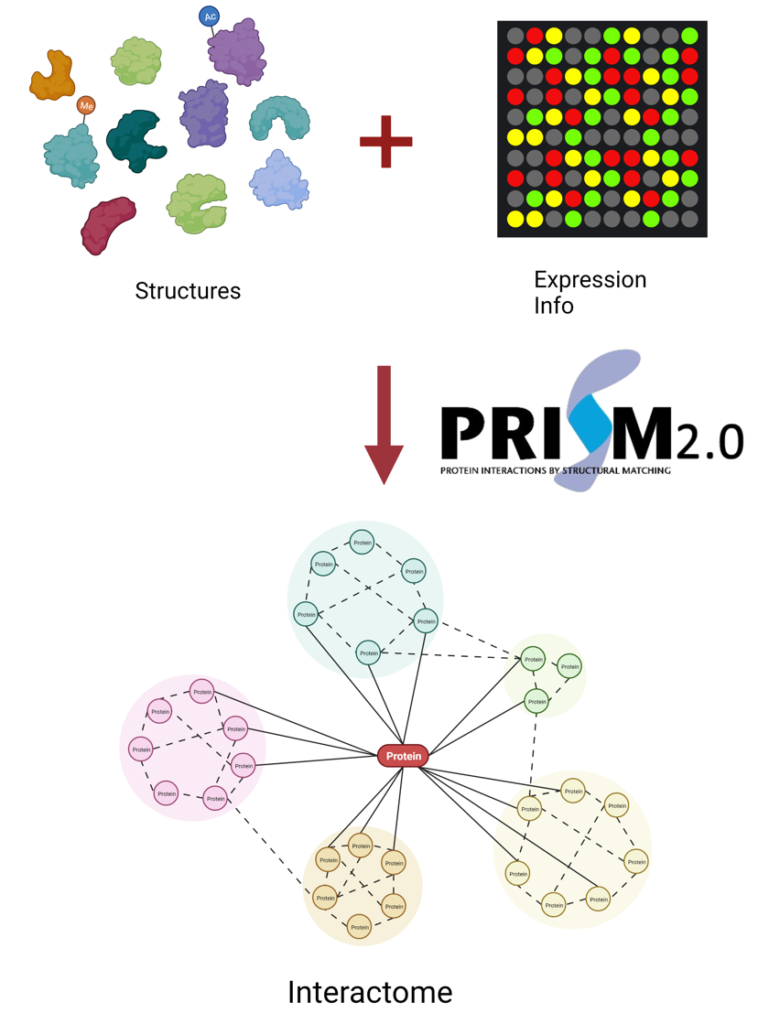
In this project, we are building the most comprehensive set of human interactome via combining available experimental and computational datasets, retaining available expression information. We will apply the PRISM algorithm to each potential interaction partner pair to build the cell type, cell cycle, and subcellular localization specific human interactome.
At the molecular level, all functions performed by cells are mediated by protein-protein or protein-molecule interactions. The complete set of interactions that can occur in an organism is called the interactome. It is of special importance to examine the interactome, which shows the relationship between proteins as a whole, to interpret protein functions and complex cellular processes, to explain the events that cause diseases, and to decipher genotype-phenotype relationships.[2]
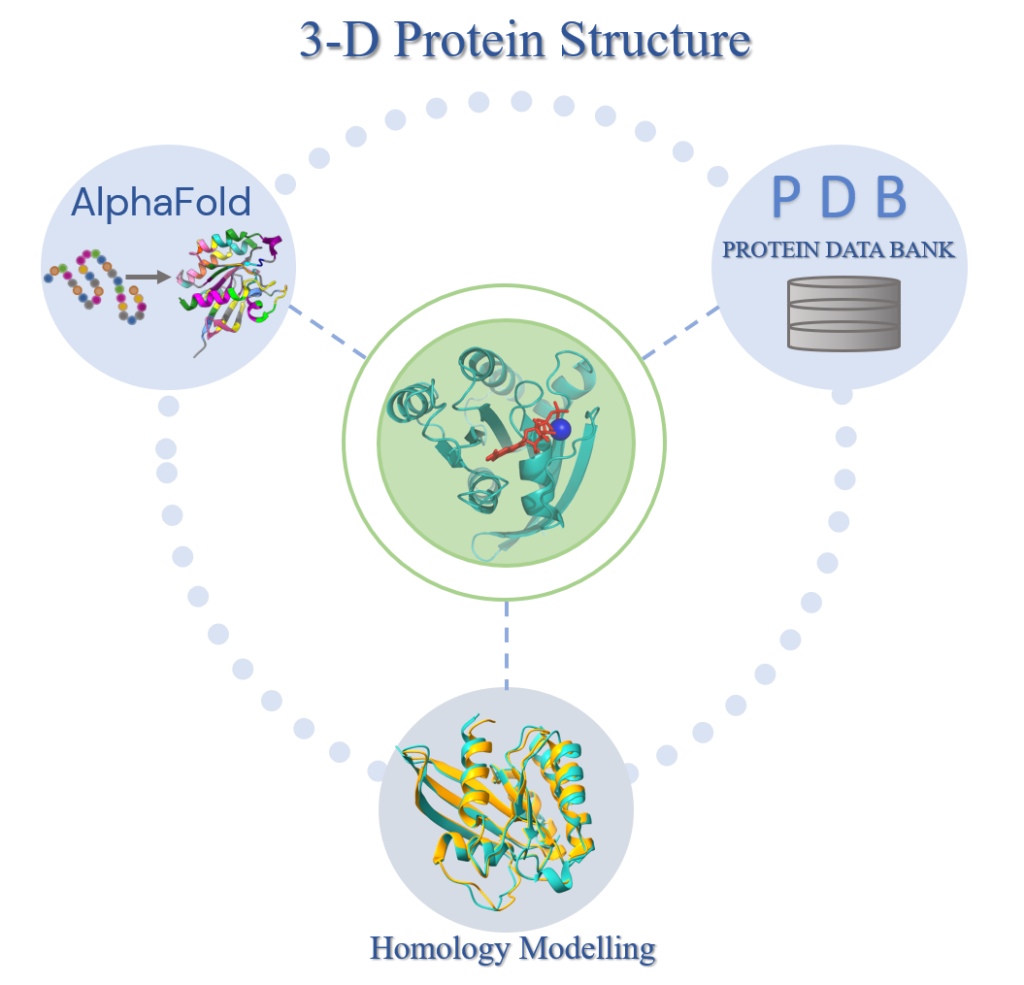
Increasing structural availability of human proteome, in combination with the advancements in protein-protein structure prediction methods paves the way to a complete human interactome. Both experimental and computational approaches have enabled us to reach to a near complete set of human proteome which can be processed with interaction prediction algorithms to obtain the interactome.
Predicting protein-protein interactions at the proteome scale, however, is a computationally challenging task. PRISM, a template-based protein-protein interaction prediction algorithm, exploit the fact that many globally different protein structures interact via similar architectural motifs.[3,4] This constitutes an efficient approach to perform a pairwise comparison of the human proteome to obtain the interactome map.
References
1. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature, (2001)
2. Keskin, O., Tuncbag, N., Gursoy, A. Predicting Protein–Protein Interactions from the Molecular to the Proteome Level. Chemical Reviews, (2016)
3. Tuncbag, N., Gursoy, A., Nussinov, R., Keskin, O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nature Protocols 6, (2011)
4. Keskin, O., Gursoy, A. Ma, B., Nussinov, R. Principles of Protein−Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chemical Reviews, (2008)
Project Members
• Kayra Kösoğlu
• Zeynep Aydın
• Fatma Cankara
• Amir Fathi
• Zeynep Abalı
• Yiğit Can Ateş